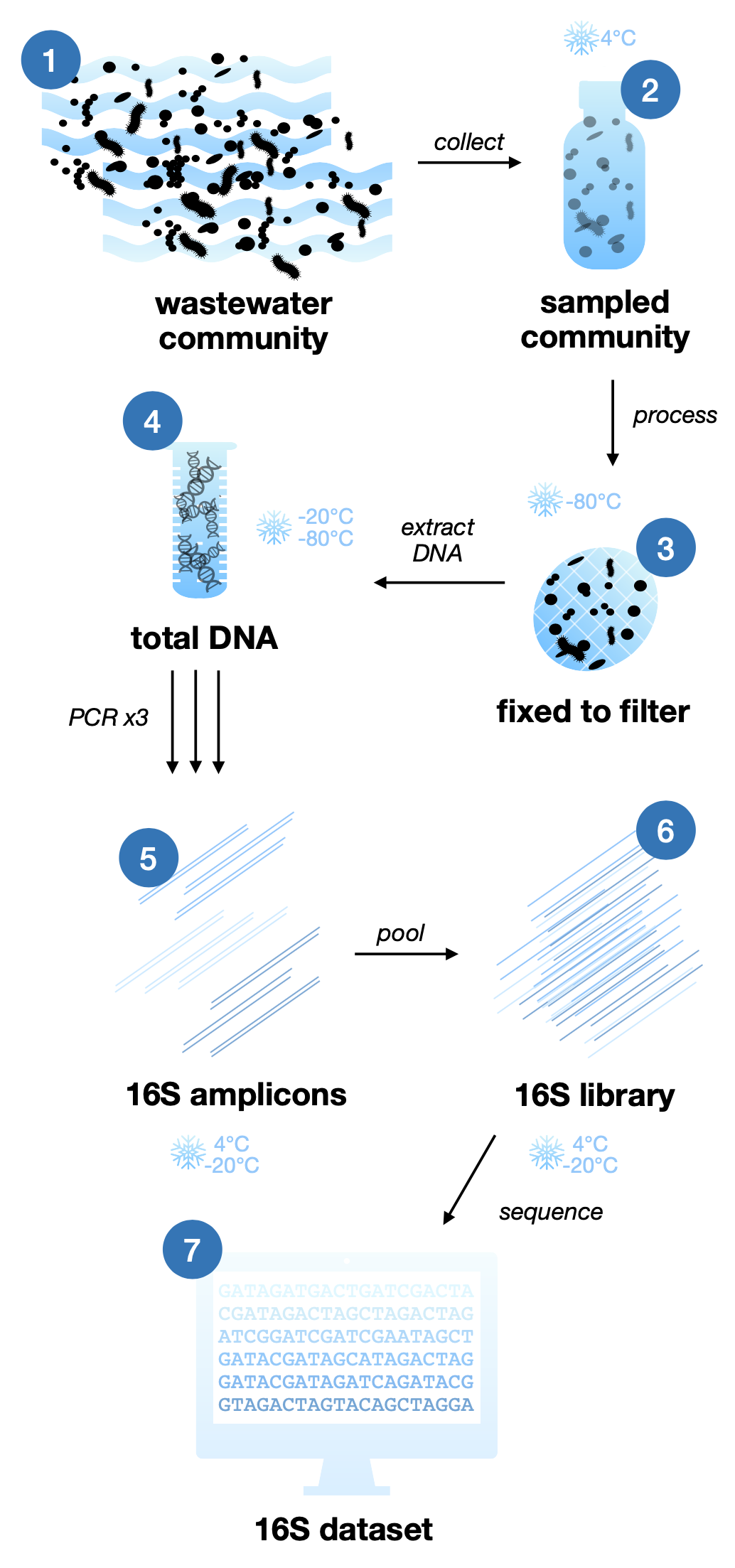
Microbial marker gene reference database for wastewater

Data availability
ASV files
Amplicon sequence variants (ASVs), or unique DNA sequences, of 16S ribosomal RNA genes from wastewater bacteria. Counts files are the number of times (reads) that ASVs occur in each sample. Taxonomy files show the taxonomic classification of ASVs from Kingdom to Species. ASV names range from ASV0001 to ASV1041, ranked from most to least abundant. FASTA sequences of ASVs whose headers include ASV ID, taxonomic assignments, read count, and read direction (R1/R2).
| Counts | GitHub | |
| Taxonomy | GitHub | |
| FASTA | GitHub |
OTU files
Operational taxonomic units (OTUs) were generated by grouping ASVs that were at least 99.5% similar. OTU names range from OTU001 to OTU681, ranked from most to least abundant. If there was no consensus in taxonomy among ASVs within an OTU, the proportion of reads belonging to that ASV is in its name. For example, OTU011 was 16 ASVs all in the genus Acidovorax, but they were mixed with defluvii (11), carolinensis (4), or were unclassified (1) to species. Among all the reads in OTU011 (5568), 67% (3719) were assigned to defluvii, while carolinensis and unclassified were 16% and 17%, respectively. Therefore, the OTU names are OTU011_67, OTU011_17, and OTU011_16.
| Counts | GitHub | |
| Taxonomy | GitHub |
Raw files, R Data, and code
Phyloseq is an R object with ASV or OTU counts, taxonomy, and sample information combined, for easy exploration in R. If you want to recreate the analysis, output files from each step (code script) are included.
| FASTQs | NCBI Short Read Archive |
| Phyloseq | ASV | OTU |
| trim residual primers | code | input & input |
| dereplicate trimmed reads | code | input & input |
| subset sewage samples | code | input & input |
| cluster ASVs to OTUs | code | input & input |
| assess taxonomy | code | input & input |
Sample set
In total, 46 wastewater treatment plant influent (raw sewage) underwent 16S rRNA gene sequencing. Samples encompass a wide range of bacterial diversity over space and time, according to previous studies (1, 2). Temporally, 24 sewage samples were collected once a month for two years from a single treatment plant. Spatially, 22 treatment plants were sampled from across the US, with southern samples from summer and northern samples from winter.
| Metadata | GitHub |
Analysis
-
Marker gene. Hypervariable and conserved regions (V1-V9) were PCR-amplified at 27F and 1492R. Unique barcodes were appended to primers to allow sequencing of all samples simultaneously (multiplex).
-
DNA sequencing. PCR amplicons were sequenced in multiplex on a PacBio Sequel II.
-
Data processing. Data files were subsetted to individual samples according to their assigned barcodes. Cutadapt was used to trim primers and barcodes from reads, DADA2 generated ASV counts and assigned taxonomy, and mothur clustered ASVs into OTUs.
Results
Common OTUs
The most common OTUs are distinct between datasets. Communities expected to have a “warm” assemblage, such as those from the South US, are very different from “cold” communities. Entire genera such as Trichococcus were completely absent from the most warm-like samples. In contrast, Pseudomonas mendocina were exlusively found in South US wastewater.
Diversity
Within- (richness) and between- (similarity) sample diversity tracks what has been shown in previous studies. Short-read V4-V5 analyses showed more within-sample diversity, however, long-read full 16S rRNA genes captured 96% of the short-read ASVs. Therefore, both short- and long-read analyses are sufficient for community analysis, but short-read (Illumina) data might better capture rare organisms, while long-read (PacBio) offers greater taxonomic resolution.
Temperature dependence
As seen previously (1, 2), wastewater temperature is a strong driver of microbial community structures. Warm-like wastewater samples cluster apart from cold ones. Further, relative abundances of OTUs fluctuate according to those temperatures.